|
|
|||
| NRAO Home > CASA > CASA Toolkit Reference Manual |
|
||
image.statistics - Function
1.1.1 Compute statistics from the image
Description
This function computes statistics from the pixel values in the image. You can
then list them and retrieve them (into a record) for further analysis.
The chunk of the image over which you evaluate the statistics is specified by an array of axis numbers (argument axes). For example, consider a 3-dimensional image for which you specify axes=[0,2]. The statistics would be computed for each XZ (axes 0 and 2) plane in the image. You could then examine those statistics as a function of the Y (axis 1) axis. Or perhaps you set axes=[2], whereupon you could examine the statistics for each Z (axis 2) profile as a function of X and Y location in the image.
Each statistic is stored in an array in one named field in the returned record. The shape of that array is that of the axes which you did not evaluate the statistics over. For example, in the second example above, we set axes=[2] and asked for statistics as a function of the remaining axes, in this case, the X and Y (axes 0 and 1) axes. The shape of each statistics array is then [nx,ny].
The names of the fields in this record are the same as the names of the statistics that you can plot:
- npts - the number of unmasked points used
- sum - the sum of the pixel values: ∑ Ii
- flux - flux or flux density, see below for details
- sumsq - the sum of the squares of the pixel values: ∑ Ii2
-
mean
-
the
mean
of
pixel
values:
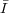 = ∑
Ii∕n
= ∑
Ii∕n
-
sigma
-
the
standard
deviation
about
the
mean:
σ2 = (∑
Ii -
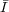 )2∕(n - 1)
)2∕(n - 1)
-
rms
-
the
root
mean
square:
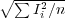
- min - minimum pixel value
- max - the maximum pixel value
- median - the median pixel value (if robust=T)
- medabsdevmed - the median of the absolute deviations from the median (if robust=T)
- quartile - the inter-quartile range (if robust=T). Find the points which are 25% largest and 75% largest (the median is 50% largest).
- q1 - The first quartile. Reported only if robust=T.
- q3 - The third quartile. Reported only if robust=T.
- blc - the absolute pixel coordinate of the bottom left corner of the bounding box of the region of interest. If ’region’ is unset, this will be the bottom left corner of the whole image.
- blcf - the formatted absolute world coordinate of the bottom left corner of the bounding box of the region of interest.
- trc - the absolute pixel coordinate of the top right corner of the bounding box of the region of interest.
- trcf - the formatted absolute world coordinate of the top right corner of the bounding box of the region of interest.
- minpos - absolute pixel coordinate of minimum pixel value
- maxpos - absolute pixel coordinate of maximum pixel value
- minposf - formatted string of the world coordinate of the minimum pixel value
- maxposf - formatted string of the world coordinate of the maximum pixel value
The last four fields only appear if you evaluate the statistics over all axes in the image. As an example, if the returned record is captured in ‘mystats’, then you could access the ‘mean’ field via print mystats[’mean’].
If there are no good points (e.g. all pixels are masked bad in the region), then the length of these fields will be 0 (e.g. len(mystats[’npts’])==0).
You have no control over which statistics are listed to the logger, you always get the same selection. You can choose to list the statistics or not (argument list).
As well as the simple (and faster to calculate) statistics like means and sums, you can also compute some robust (quantile-like) statistics. Currently these are the median, median absolute deviations from the median, the first and third quartiles, and the inner-quartile range. Because these are computationally expensive, they are only computed if robust=True.
Note that if the axes are set to all of the axes in the image (which is the default) there is just one value per statistic.
You have control over which pixels are included in the statistics computations via the includepix and excludepix arguments. These vectors specify a range of pixel values for which pixels are either included or excluded. They are mutually exclusive; you can specify one or the other, but not both. If you only give one value for either of these, say includepix=b, then this is interpreted as includepix=[-abs(b),abs(b)].
This function generates a ‘storage’ lattice, into which the statistics are written. It is only regenerated when necessary. For example, if you run the function twice with identical arguments, the statistics will be directly retrieved from the storage lattice the second time. However, you can force regeneration of the storage image if you set force=T. The storage medium is either in memory or on disk, depending upon its size. You can force it to disk if you set disk=T, otherwise it decides for itself.
ALGORITHMS
Several types of statistical algorithms are supported:
* classic: This is the familiar algorithm, in which all unmasked pixels, subject to any specified pixel ranges, are used. One may choose one of two methods, which vary only by performance, for computing classic statistics, via the clmethod parameter. The ”tiled” method is the old method and is fastest in cases where there are a large number of individual sets of statistics to be computed and a small number of data points per set. This can occur when one sets the axes parameter, which causes several individual sets of statistics to be computed. The ”framework” method uses the new statistics framework to compute statistics. This method is fastest in the regime where one has a small number of individual sets of statistics to calculate, and each set has a large number of points. For example, this method is fastest when computing statistics over an entire image in one go (no axes specified). A third option, ”auto”, chooses which method to use by predicting which be faster based on the number of pixels in the image and the choice of the axes parameter.
* fit-half: This algorithm calculates statistics on a dataset created from real and virtual pixel values. The real values are determined by the input parameters center and lside. The parameter center tells the algorithm where the center value of the combined real+virtual dataset should be. Options are the mean or the median of the input image’s pixel values, or at zero. The lside parameter tells the algorithm on which side of this center the real pixel values are located. True indicates that the real pixel values to be used are ¡= center. False indicates the real pixel values to be used are ¿= center. The virtual part of the dataset is then created by reflecting all the real values through the center value, to create a perfectly symmetric dataset composed of a real and a virtual component. Statistics are then calculated on this resultant dataset. These two parameters are ignored if algorithm is not ”fit-half”. Because the maximum value is virtual if lside is True and the minimum value is virtual if lside is False, the value of the maximum position (if lside=True) or minimum position (if lside=False) is not reported in the returned record.
* hinges-fences: This algorithm calculates statistics by including data in a range between Q1 - f*D and Q3 + f*D, inclusive, where Q1 is the first quartile of the distribution of unmasked data, subject to any specified pixel ranges, Q3 is the third quartile, D = Q3 - Q1 (the inner quartile range), and f is the user-specified fence factor. Negative values of f indicate that the full distribution is to be used (ie, the classic algorithm is used). Sufficiently large values of f will also be equivalent to using the classic algorithm. For f = 0, only data in the inner quartile range is used for computing statistics. The value of fence is silently ignored if algortihm is not ”hinges-fences”.
* chauvenet: The idea behind this algorithm is to eliminate outliers based on a maximum z-score value. A z-score is the number of standard deviations a point is from the mean of a distribution. This method thus is meant to be used for (nearly) normal distributions. In general, this is an iterative process, with successive iterations discarding additional outliers as the remaining points become closer to forming a normal distribution. Iterating stops when no additional points lie beyond the specified zscore value, or, if zscore is negative, when Chauvenet’s criterion is met (see below). The parameter maxiter can be set to a non-negative value to prematurely abort this iterative process. When verbose=True, the ”N iter” column in the table that is logged represents the number of iterations that were executed.
Chauvenet’s criterion allows the target z-score to decrease as the number of points in the distribution decreases on subsequent iterations. Essentially, the criterion is that the probability of having one point in a normal distribution at a maximum z-score of z_max must be at least 0.5. z_max is therefore a function of (only) the number of points in the distrbution and is given by
npts = 0.5/erfc(z_max/sqrt(2))
where erfc() is the complementary error function. As iterating proceeds, the number of remaining points decreases as outliers are discarded, and so z_max likewise decreases. Convergence occurs when all remaining points fall within a z-score of z_max. Below is an illustrative table of z_max values and their corresponding npts values. For example, it is likely that there will be a 5-sigma ”noise bump” in a perfectly noisy image with one million independent elements.
z_max npts 1.0 1 1.5 3 2.0 10 2.5 40 3.0 185 3.5 1,074 4.0 7,893 4.5 73,579 5.0 872,138 5.5 13,165,126 6.0 253,398,672 6.5 6,225,098,696 7.0 195,341,107,722
NOTES ON FLUX DENSITIES AND FLUXES
Fluxes and flux densities are not computed if any of the following conditions is met:
1. The image does not have a direction coordinate 2. The image does not have a intensity-like brightness unit. Examples of such units are Jy/beam (in which case the image must also have a beam) and K. 3. There are no direction axes in the cursor axes that are used. 4. If the (specified region of the) image has a non-degenerate spectral axis, and the image has a tablular spectral axis (axis with varying increments) 5. Any axis that is not a direction nor a spectral axis that is included in the cursor axes is not degenerate within in the specified region
Note that condition 4 may be removed in the future.
In cases where none of the above conditions is met, the flux density(ies) (intensities integrated over direction planes) will be computed if any of the following conditions are met:
1. The image has no spectral coordinate 2. The cursor axes do not include the spectral axis 3. The spectral axis in the chosen region is degenerate
In the case where there is a nondegenerate spectral axis that is included in the cursor axes, the flux (flux density integrated over spectral planes) will be computed. In this case, the spectral portion of the flux unit will be the velocity unit of the spectral coordinate if it has one (eg, if the brightness unit is Jy/beam and the velocity unit is km/s, the flux will have units of Jy.km/s). If not, the spectral portion of the flux unit will be the frequency unit of the spectral axis (eg, if the brightness unit is K and the frequency unit is Hz, the resulting flux unit will be K.arcsec2.Hz).
In both cases of flux density or flux being computed, the resulting numerical value is assigned to the ”flux” key in the output dictionary.
Arguments
| Inputs |
| ||
| axes |
| List of axes to evaluate statistics over. Default is all axes. | |
| allowed: | intArray |
|
| Default: | -1 | |
| region |
| Region selection. Default is to use the full image.
| |
| allowed: | any |
|
| Default: | variant
|
|
| mask |
| Mask to use. Default is none. | |
| allowed: | any |
|
| Default: | variant
|
|
| includepix |
| Range of pixel values to include. Vector of 1 or 2 doubles.
Default is to include all pixels.
| |
| allowed: | doubleArray |
|
| Default: | -1 | |
| excludepix |
| Range of pixel values to exclude. Vector of 1 or 2 doubles.
Default is exclude no pixels.
| |
| allowed: | doubleArray |
|
| Default: | -1 | |
| list |
| If True print bounding box and statistics to logger.
| |
| allowed: | bool | |
| Default: | false |
|
| force |
| If T then force the stored statistical accumulations to be
regenerated
| |
| allowed: | bool | |
| Default: | false |
|
| disk |
| If T then force the storage image to disk
| |
| allowed: | bool | |
| Default: | false |
|
| robust |
| If T then compute robust statistics as well
| |
| allowed: | bool | |
| Default: | false |
|
| verbose |
| If T then log statistics
| |
| allowed: | bool | |
| Default: | false |
|
| stretch |
| Stretch the mask if necessary and possible? Default False
| |
| allowed: | bool | |
| Default: | false |
|
| logfile |
| Name of file to which to write statistics.
| |
| allowed: | string |
|
| Default: |
| |
| append |
| Append results to logfile? Logfile must be specified.
Default is to append. False means overwrite existing file
if it exists.
| |
| allowed: | bool |
|
| Default: | true |
|
| algorithm |
| Algorithm to use. Supported values are ”chauvenet”,
”classic”, ”fit-half”, and ”hinges-fences”. Minimum
match is supported.
| |
| allowed: | string |
|
| Default: | classic |
|
| fence |
| Fence value for hinges-fences. A negative value means
use the entire data set (ie default to the ”classic”
algorithm). Ignored if algorithm is not ”hinges-fences”.
| |
| allowed: | double |
|
| Default: | -1 |
|
| center |
| Center to use for fit-half. Valid choices are ”mean”,
”median”, and ”zero”. Ignored if algorithm is not
”fit-half”.
| |
| allowed: | string |
|
| Default: | mean |
|
| lside |
| For fit-half, real data are <=; center? If false, real data
are >= center. Ignored if algorithm is not ”fit-half”.
| |
| allowed: | bool |
|
| Default: | true |
|
| zscore |
| For chauvenet, this is the target maximum number of
standard deviations data may have to be included. If
negative, use Chauvenet’s criterion. Ignored if algorithm
is not ”chauvenet”.
| |
| allowed: | double |
|
| Default: | -1 |
|
| maxiter |
| For chauvenet, this is the maximum number of iterations
to attempt. Iterating will stop when either this limit is
reached, or the zscore criterion is met. If negative, iterate
until the zscore criterion is met. Ignored if algortihm is
not ”chauvenet”.
| |
| allowed: | int |
|
| Default: | -1 |
|
| clmethod |
| Method to use for calculating classical
statistics. Supported methods are ”auto”, ”tiled”, and
”framework”. Ignored if algorithm is not ”classic”.
| |
| allowed: | string |
|
| Default: | auto |
|
record
Example
"""
#
print "\t----\t statistics Ex 1 \t----"
ia.maketestimage()
ia.statistics()
ia.close()
#
# evaluate statistics for each spectral plane in an ra x dec x frequency image
ia.fromshape("", [20,30,40])
# give pixels non-zero values
ia.addnoise()
# These are the display axes, the calculation of statistics occurs
# for each (hyper)plane along axes not listed in the axes parameter,
# in this case axis 2 (the frequency axis)
# display the rms for each frequency plane (your mileage will vary with
# the values).
stats = ia.statistics(axes=[0,1])
stats["rms"]
Out[10]:
array([ 0.99576014, 1.03813124, 0.97749186, 0.97587883, 1.04189885,
1.03784776, 1.03371549, 1.03153074, 1.00841606, 0.947155 ,
0.97335404, 0.94389403, 1.0010221 , 0.97151822, 1.03942156,
1.01158476, 0.96957082, 1.04212773, 1.00589049, 0.98696715,
1.00451481, 1.02307892, 1.03102005, 0.97334671, 0.95209879,
1.02088714, 0.96999902, 0.98661619, 1.01039267, 0.96842754,
0.99464947, 1.01536798, 1.02466023, 0.96956468, 0.98090756,
0.9835844 , 0.95698935, 1.05487967, 0.99846411, 0.99634868])
"""
In this example, we ask to see statistics evaluated over the
entire image.
Example
"""
#
print "\t----\t statistics Ex 2 \t----"
ia.maketestimage()
stats = ia.statistics(axes=[1],plotstats=["sigma","rms"],
includepix=[0,100],list=False)
#
"""
In this example, let us assume the image has 2 dimensions. We want
the standard deviation about the mean and the rms of Y (axes 1) for
pixels with values in the range 0 to 100 as a function of the X-axis
location. The statistics are not listed to the logger but are saved
in the record {\stfaf ’stats’}.
__________________________________________________________________
More information about CASA may be found at the
CASA web page
Copyright © 2016 Associated Universities Inc., Washington, D.C.
This code is available under the terms of the GNU General Public Lincense
Home |
Contact Us |
Directories |
Site Map |
Help |
Privacy Policy |
Search
